Introduction
Overall, my research aims to understand how environmental factors, like wild fires, influence biodiversity. One of the most used databases for measuring biodiversity is the Global Biodiversity Information Facility GBIF. This is post is a brief introduction of how to use GBIF, using the example of mapping a few conifer species in California.
Load the libraries
library(rgbif)
library(tidyverse)
library(data.table)
library(maps)
library(leaflet)Part 1 -Mapping one species
Using the R package function occ_data() request data from the GBIF database with the species name, selecting the observations that have a coordinate, and limiting the query to 10000 observations.
pinus_balfouriana <- occ_data(scientificName = "Pinus balfouriana", hasCoordinate = TRUE, limit = 10000)Inspect the data structure.
summary(pinus_balfouriana)## Length Class Mode
## meta 4 -none- list
## data 134 tbl_df listThere are many different groups that contribute data to GBIF. Make sure you cite them accordingly so we can continue to have a great stream of species occurrence data. As a a few examples this species has currated research grade observations from iNaturalist and many from the Humboldt State University.
head(gbif_citation(pinus_balfouriana), 2)## [[1]]
## <<rgbif citation>>
## Citation: iNaturalist contributors, iNaturalist (2022). iNaturalist
## Research-grade Observations. iNaturalist.org. Occurrence dataset
## https://doi.org/10.15468/ab3s5x accessed via GBIF.org on 2022-04-05..
## Accessed from R via rgbif (https://github.com/ropensci/rgbif) on
## 2022-04-05
## Rights: http://creativecommons.org/licenses/by-nc/4.0/legalcode
##
## [[2]]
## <<rgbif citation>>
## Citation: Humboldt State University (2022). HSC - Humboldt State University
## Vascular Plant Herbarium. Occurrence dataset
## https://doi.org/10.15468/qguk7r accessed via GBIF.org on 2022-04-05..
## Accessed from R via rgbif (https://github.com/ropensci/rgbif) on
## 2022-04-05
## Rights: http://creativecommons.org/licenses/by-nc/4.0/legalcodeTake a look at what types of data are collected.
head(names(pinus_balfouriana$data))## [1] "key" "scientificName" "decimalLatitude" "decimalLongitude"
## [5] "issues" "datasetKey"The default GBIF query returns 126 columns of data. We do not have time to go through all of them for a single post so I will subset some important ones for some exploratory plotting purposes. Here I subset the Latitude and Longitude coordinates that we will use for mapping, if the species does occur (important for modeling in future posts), how uncertain the observation is in meters, and the references for the observation.
pinus_balfouriana_coords <- pinus_balfouriana$data[ , c("decimalLongitude", "decimalLatitude",
"occurrenceStatus", "coordinateUncertaintyInMeters", "references")]# check out how the data is structured
head(pinus_balfouriana_coords)## # A tibble: 6 × 5
## decimalLongitude decimalLatitude occurrenceStatus coordinateUncert… references
## <dbl> <dbl> <chr> <dbl> <chr>
## 1 -118. 36.6 PRESENT 1742 https://w…
## 2 -118. 36.8 PRESENT 130 https://w…
## 3 -118. 36.5 PRESENT NA https://w…
## 4 -118. 36.5 PRESENT NA https://w…
## 5 -118. 36.4 PRESENT NA https://w…
## 6 -118. 36.4 PRESENT NA https://w…summary(pinus_balfouriana_coords)## decimalLongitude decimalLatitude occurrenceStatus
## Min. :-123.21 Min. :35.92 Length:417
## 1st Qu.:-122.53 1st Qu.:36.49 Class :character
## Median :-118.56 Median :36.77 Mode :character
## Mean :-119.26 Mean :38.14
## 3rd Qu.:-118.32 3rd Qu.:40.95
## Max. : 11.45 Max. :50.83
##
## coordinateUncertaintyInMeters references
## Min. : 2 Length:417
## 1st Qu.: 10 Class :character
## Median : 201 Mode :character
## Mean : 2479
## 3rd Qu.: 802
## Max. :28534
## NA's :152# pinus_balfouriana_coords$decimalLongitude # remove this row of bad data
pinus_balfouriana_coords <- slice(pinus_balfouriana_coords, -(278))# two map functions... be clear! There are a few map functions with these different libraries loaded.
#?map()
maps::map(database = "state", region = "california")
points(pinus_balfouriana_coords[ , c("decimalLongitude", "decimalLatitude")], pch = ".", col = "red", cex = 3)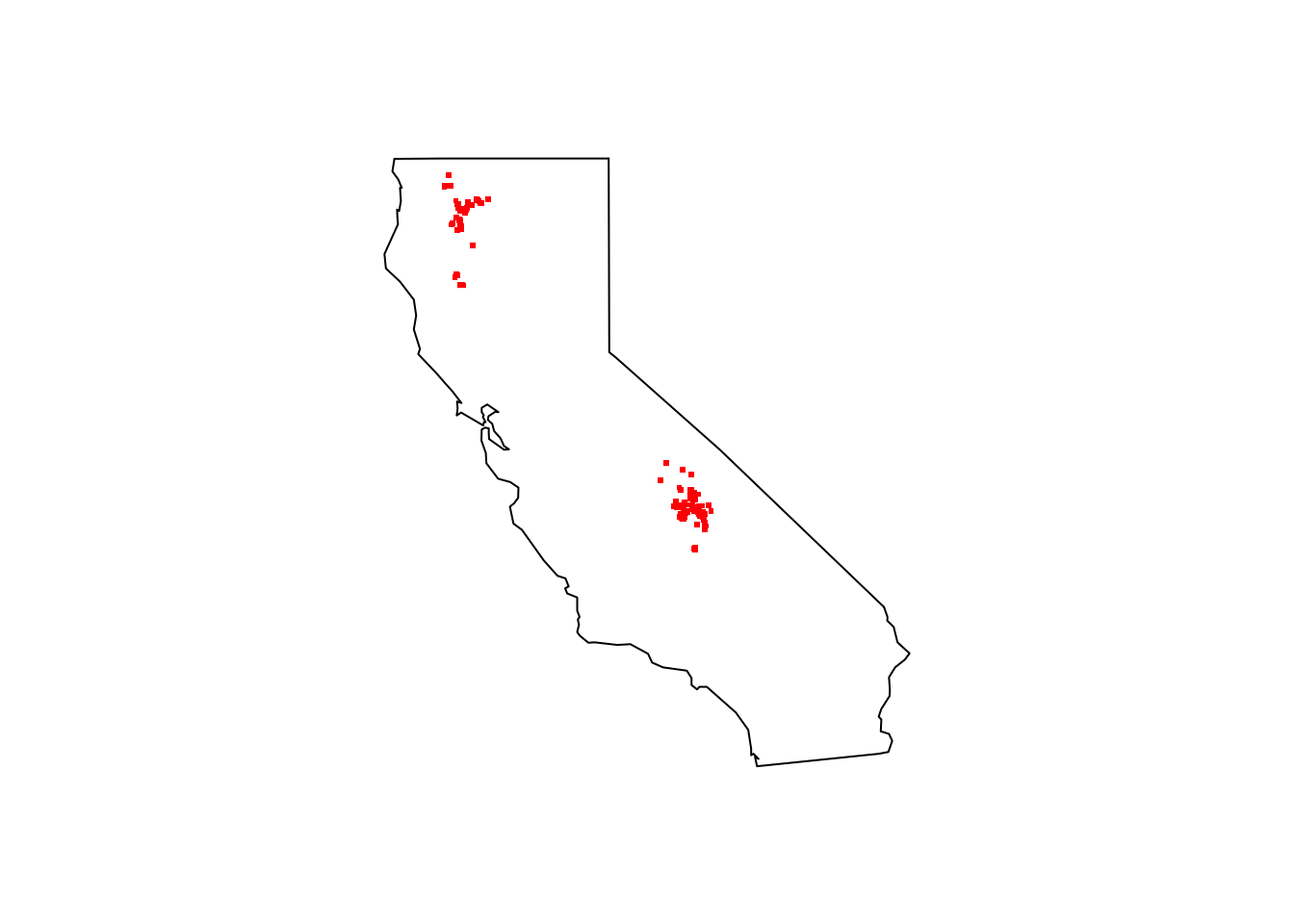
Subset our search to only Northern California
mm_geometry <- paste('POLYGON((-124.4323 42.0021, -121.5045 42.0021, -121.5045 40.194, -124.4323 40.194, -124.4323 42.0021))')pinus_balfouriana_NC <- occ_data(scientificName = "Pinus balfouriana", hasCoordinate = TRUE, limit = 10000,
geometry = mm_geometry )
head(pinus_balfouriana_NC)## $meta
## $meta$offset
## [1] 0
##
## $meta$limit
## [1] 300
##
## $meta$endOfRecords
## [1] TRUE
##
## $meta$count
## [1] 121
##
##
## $data
## # A tibble: 121 × 122
## key scientificName decimalLatitude decimalLongitude issues datasetKey
## <chr> <chr> <dbl> <dbl> <chr> <chr>
## 1 3455305209 Pinus balfouri… 41.3 -123. cdrou… 50c9509d-…
## 2 3328047476 Pinus balfouri… 41.3 -122. cdrou… 50c9509d-…
## 3 3456635831 Pinus balfouri… 40.9 -123. cdrou… 50c9509d-…
## 4 3456682356 Pinus balfouri… 41.2 -123. cdrou… 50c9509d-…
## 5 2620109834 Pinus balfouri… 40.7 -123. cdrou… 50c9509d-…
## 6 2814362317 Pinus balfouri… 41.2 -123. cdrou… 50c9509d-…
## 7 2850958731 Pinus balfouri… 41.3 -122. cdrou… 50c9509d-…
## 8 2862286454 Pinus balfouri… 41.3 -123. cdrou… 50c9509d-…
## 9 3067629037 Pinus balfouri… 40.9 -123. cdrou… 50c9509d-…
## 10 3384272954 Pinus balfouri… 40.9 -123. cdrou… 50c9509d-…
## # … with 111 more rows, and 116 more variables: publishingOrgKey <chr>,
## # installationKey <chr>, publishingCountry <chr>, protocol <chr>,
## # lastCrawled <chr>, lastParsed <chr>, crawlId <int>,
## # hostingOrganizationKey <chr>, basisOfRecord <chr>, occurrenceStatus <chr>,
## # taxonKey <int>, kingdomKey <int>, phylumKey <int>, classKey <int>,
## # orderKey <int>, familyKey <int>, genusKey <int>, speciesKey <int>,
## # acceptedTaxonKey <int>, acceptedScientificName <chr>, kingdom <chr>, …pinus_balfouriana_NC_coords <- pinus_balfouriana_NC$data[ , c("decimalLongitude", "decimalLatitude",
"individualCount", "occurrenceStatus", "coordinateUncertaintyInMeters",
"institutionCode", "references")]Plot the data in a few different ways to see if there is anything strange.
plot(pinus_balfouriana_NC_coords$decimalLongitude, pinus_balfouriana_NC_coords$decimalLatitude) # examine the data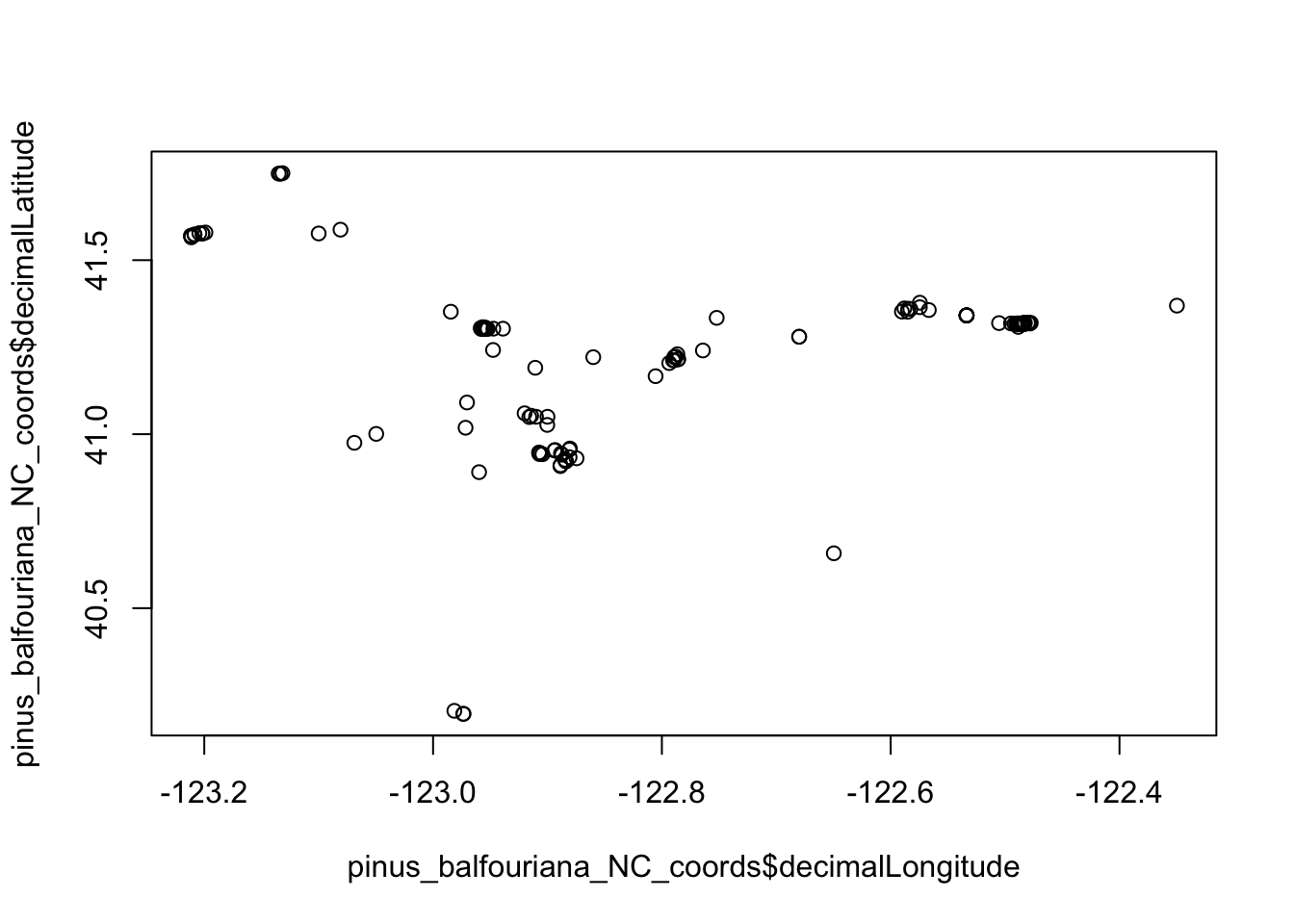
pinus_balfouriana_NC_coords %>% leaflet() %>% addTiles() %>%
addMarkers(~decimalLongitude, ~decimalLatitude)Part 2- Map Multiple species
First test the workflow with only a few species then do entire batch.
# The test is commented out. Uncomment to test first.
# mm_species <- c("Pinus balfouriana", "pinus albicaulis", "pinus monticola") # uncomment to run small test version
# Entire miracle mile species set
mm_species <- c("Pinus balfouriana", "pinus albicaulis", "pinus monticola", "pinus jeffreyi", "pinus ponderosa", "pinus contorta", "pinus lambertiana", "abies concolor", "abies magnifica", "abies lasiocarpa", "picea engelmannii", "picea breweriana", "tsuga mertensiana", "pseudotsuga menziesii", "taxus brevifolia", "calocedrus decurrens", "juniperus communis", "juniperus occidentalis")
mm_all <- occ_data(scientificName = mm_species, hasCoordinate = TRUE, limit = 10000,
geometry = mm_geometry)
summary(mm_all)## Length Class Mode
## Pinus balfouriana 2 -none- list
## pinus albicaulis 2 -none- list
## pinus monticola 2 -none- list
## pinus jeffreyi 2 -none- list
## pinus ponderosa 2 -none- list
## pinus contorta 2 -none- list
## pinus lambertiana 2 -none- list
## abies concolor 2 -none- list
## abies magnifica 2 -none- list
## abies lasiocarpa 2 -none- list
## picea engelmannii 2 -none- list
## picea breweriana 2 -none- list
## tsuga mertensiana 2 -none- list
## pseudotsuga menziesii 2 -none- list
## taxus brevifolia 2 -none- list
## calocedrus decurrens 2 -none- list
## juniperus communis 2 -none- list
## juniperus occidentalis 2 -none- listmm_species_coords_list <- vector("list", length(mm_species))
names(mm_species_coords_list) <- mm_species
for (x in mm_species) {
coords <- mm_all[[x]]$data[ , c("decimalLongitude", "decimalLatitude", "occurrenceStatus", "coordinateUncertaintyInMeters", "institutionCode", "references")]
mm_species_coords_list[[x]] <- data.frame(species = x, coords)
}Using the rbindlist() function from the data.frame package to take all of the species observations from a list to a large data.frame. The columns are the species name, the latitude and longitude coordinates, whether or not there was an observation, if there is any uncertainty about how accurate the GPS coordinate was, what platform the observation was made on, and the specific reference for the observation. Make sure you cite the references so we can keep these rich data streams coming!
tree_df <- rbindlist(mm_species_coords_list, fill = T)
dim(tree_df)## [1] 5637 7head(tree_df)## species decimalLongitude decimalLatitude occurrenceStatus
## 1: Pinus balfouriana -122.7521 41.33424 PRESENT
## 2: Pinus balfouriana -122.4852 41.31701 PRESENT
## 3: Pinus balfouriana -122.9044 40.94235 PRESENT
## 4: Pinus balfouriana -122.9107 41.19077 PRESENT
## 5: Pinus balfouriana -122.6497 40.65773 PRESENT
## 6: Pinus balfouriana -122.7857 41.21531 PRESENT
## coordinateUncertaintyInMeters institutionCode
## 1: 27816 iNaturalist
## 2: NA iNaturalist
## 3: 77 iNaturalist
## 4: 5 iNaturalist
## 5: 7880 iNaturalist
## 6: 5 iNaturalist
## references
## 1: https://www.inaturalist.org/observations/81808167
## 2: https://www.inaturalist.org/observations/87043552
## 3: https://www.inaturalist.org/observations/86123914
## 4: https://www.inaturalist.org/observations/85415690
## 5: https://www.inaturalist.org/observations/44672814
## 6: https://www.inaturalist.org/observations/51134921Just take a quick look at the raw observations plotted by latitude and longitude.
Plot all of the species on the California map.
maps::map(database = "state", region = "california")
points(tree_df[ , c("decimalLongitude", "decimalLatitude")], pch = ".", col = "blue", cex = 3)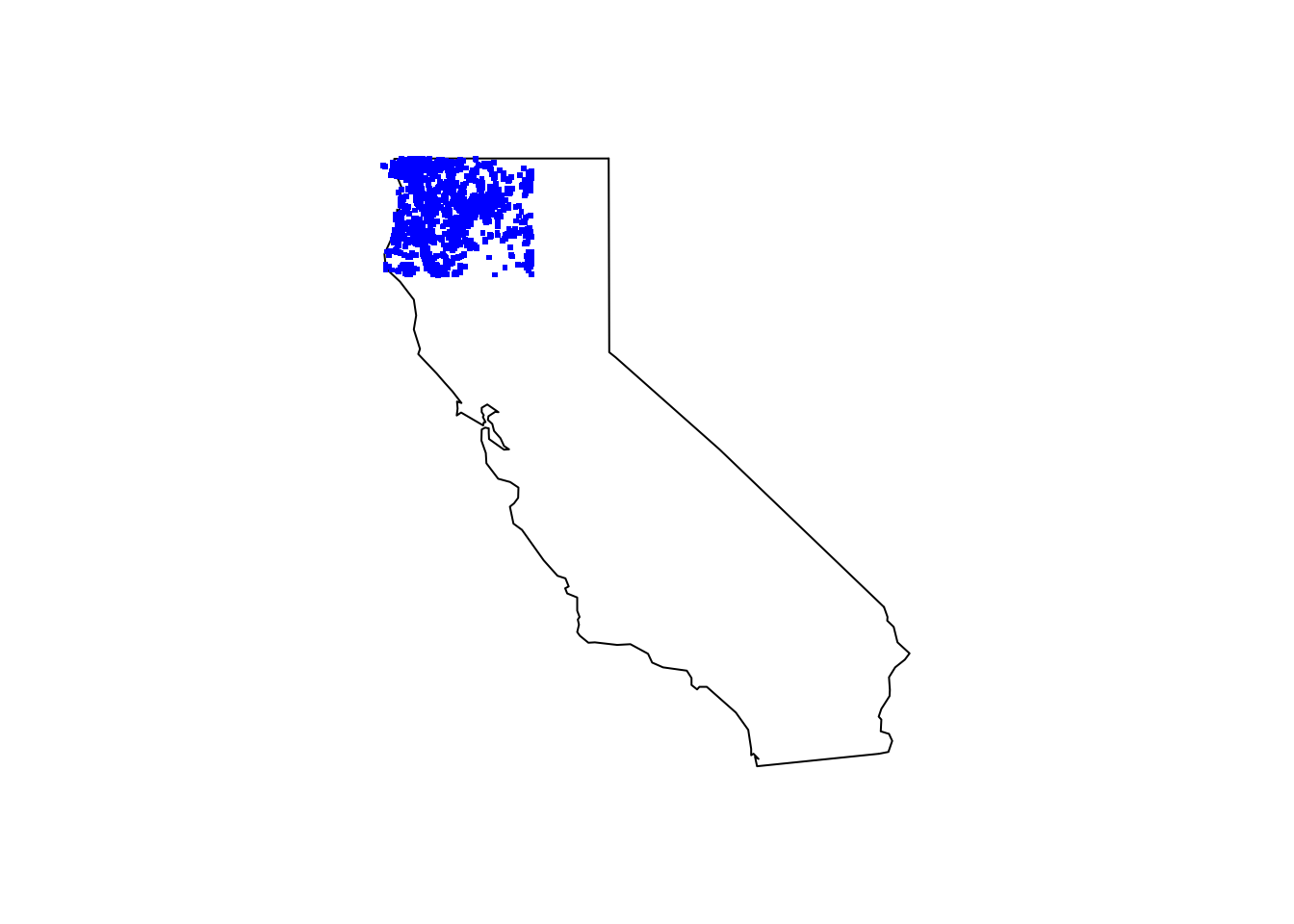
Plot all of the species using ggplot in case you want to visualize the species with something a bit fancier.
mm_species_plot1 <- ggplot(tree_df, aes(x=decimalLongitude, y = decimalLatitude, color = species)) +
geom_point() + labs(color = "Species", title = "MM Zone")
mm_species_plot1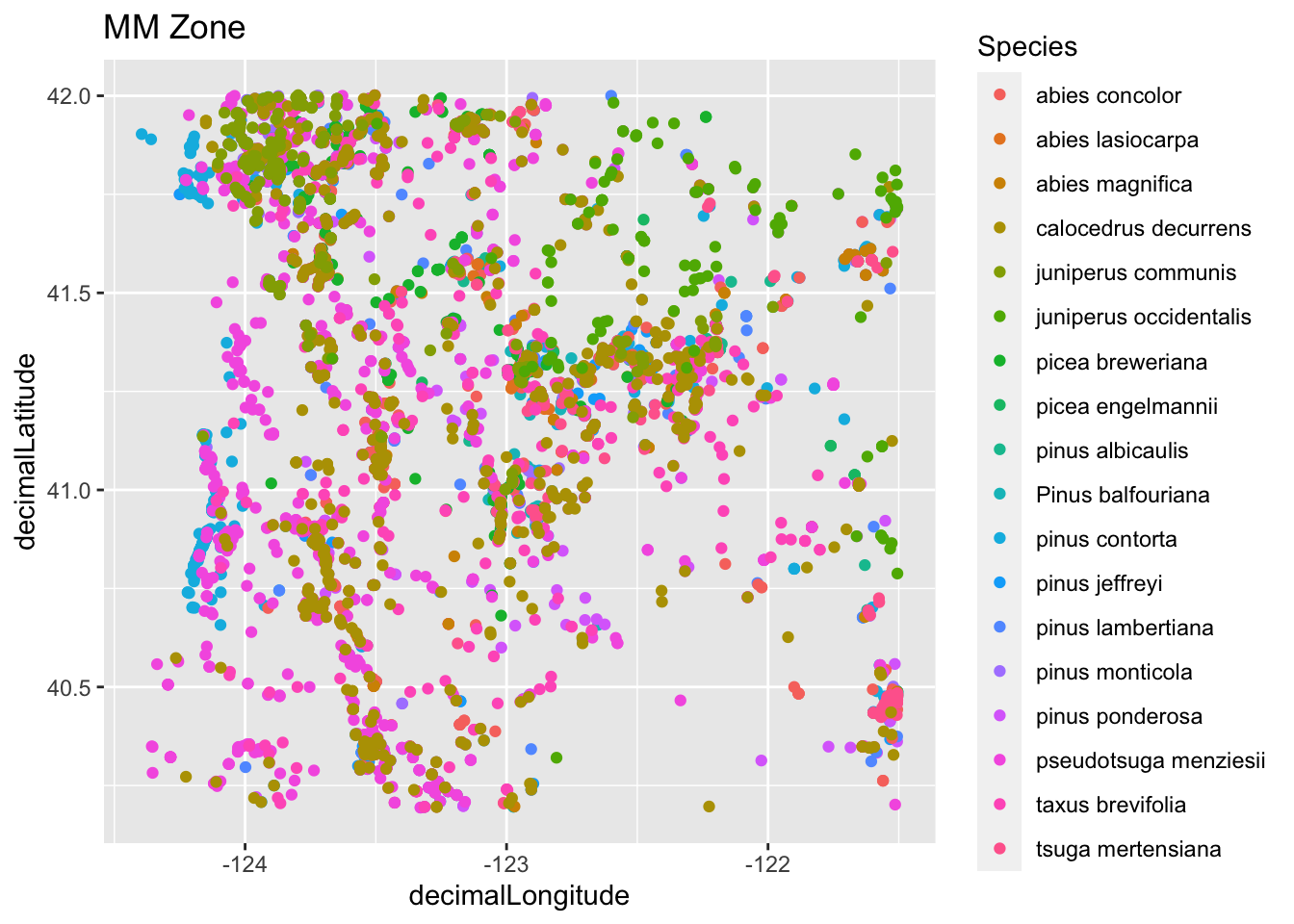
There is a lot more you can do for GBIF, but these notes should help for the purpose of mapping species occurrence. Now that the data is somewhat organized we can start doing some proper data cleaning and exploratory data analysis in a future post.