
Spring is in full swing in the higher elevations around Mount Shasta and the surrounding ranges. I am always struck by the large patches of Cobra Lilies that grow along nutrient poor seeps in alpine meadows. I nearly always stop to take some photos and take in all the cool colors and shapes when out on training runs or hikes. I have made many observations of these over the past few years, but here is a quick post to pull out observations from GBIF.
Load the libraries.
library(tidyverse)
library(rgbif)
library(gpx)Make the large Northern California polygon to look for observations in GBIF.
norcal_geometry <- paste('POLYGON((-124.4323 42.0021, -121.5045 42.0021, -121.5045 40.194, -124.4323 40.194, -124.4323 42.0021))')
cobra_lily <- occ_data(scientificName = "darlingtonia californica", hasCoordinate = TRUE, limit = 1000,
geometry = norcal_geometry)
# length(cobra_lily)
head(cobra_lily)## $meta
## $meta$offset
## [1] 900
##
## $meta$limit
## [1] 100
##
## $meta$endOfRecords
## [1] FALSE
##
## $meta$count
## [1] 1129
##
##
## $data
## # A tibble: 1,000 × 125
## key scientificName decimalLatitude decimalLongitude issues datasetKey
## <chr> <chr> <dbl> <dbl> <chr> <chr>
## 1 3499352919 Darlingtonia c… 41.9 -124. cdrou… 50c9509d-…
## 2 3499475143 Darlingtonia c… 41.9 -124. cdrou… 50c9509d-…
## 3 3499606799 Darlingtonia c… 41.8 -124. cdrou… 50c9509d-…
## 4 3499614932 Darlingtonia c… 41.9 -124. cdrou… 50c9509d-…
## 5 3499614940 Darlingtonia c… 42.0 -124. cdrou… 50c9509d-…
## 6 3499647846 Darlingtonia c… 41.9 -124. cdrou… 50c9509d-…
## 7 3698398122 Darlingtonia c… 41.9 -124. cdrou… 50c9509d-…
## 8 3743185922 Darlingtonia c… 41.9 -124. cdrou… 50c9509d-…
## 9 3746965657 Darlingtonia c… 41.9 -124. cdrou… 50c9509d-…
## 10 3747115575 Darlingtonia c… 42.0 -124. cdrou… 50c9509d-…
## # … with 990 more rows, and 119 more variables: publishingOrgKey <chr>,
## # installationKey <chr>, publishingCountry <chr>, protocol <chr>,
## # lastCrawled <chr>, lastParsed <chr>, crawlId <int>,
## # hostingOrganizationKey <chr>, basisOfRecord <chr>, occurrenceStatus <chr>,
## # taxonKey <int>, kingdomKey <int>, phylumKey <int>, classKey <int>,
## # orderKey <int>, familyKey <int>, genusKey <int>, speciesKey <int>,
## # acceptedTaxonKey <int>, acceptedScientificName <chr>, kingdom <chr>, …Quick map of observations.
cobra_lily_coords <- cobra_lily$data[ , c("decimalLongitude", "decimalLatitude", "occurrenceStatus", "coordinateUncertaintyInMeters", "institutionCode", "references")]
maps::map(database = "state", region = "california")
points(cobra_lily_coords[ , c("decimalLongitude", "decimalLatitude")], pch = ".", col = "red", cex = 3)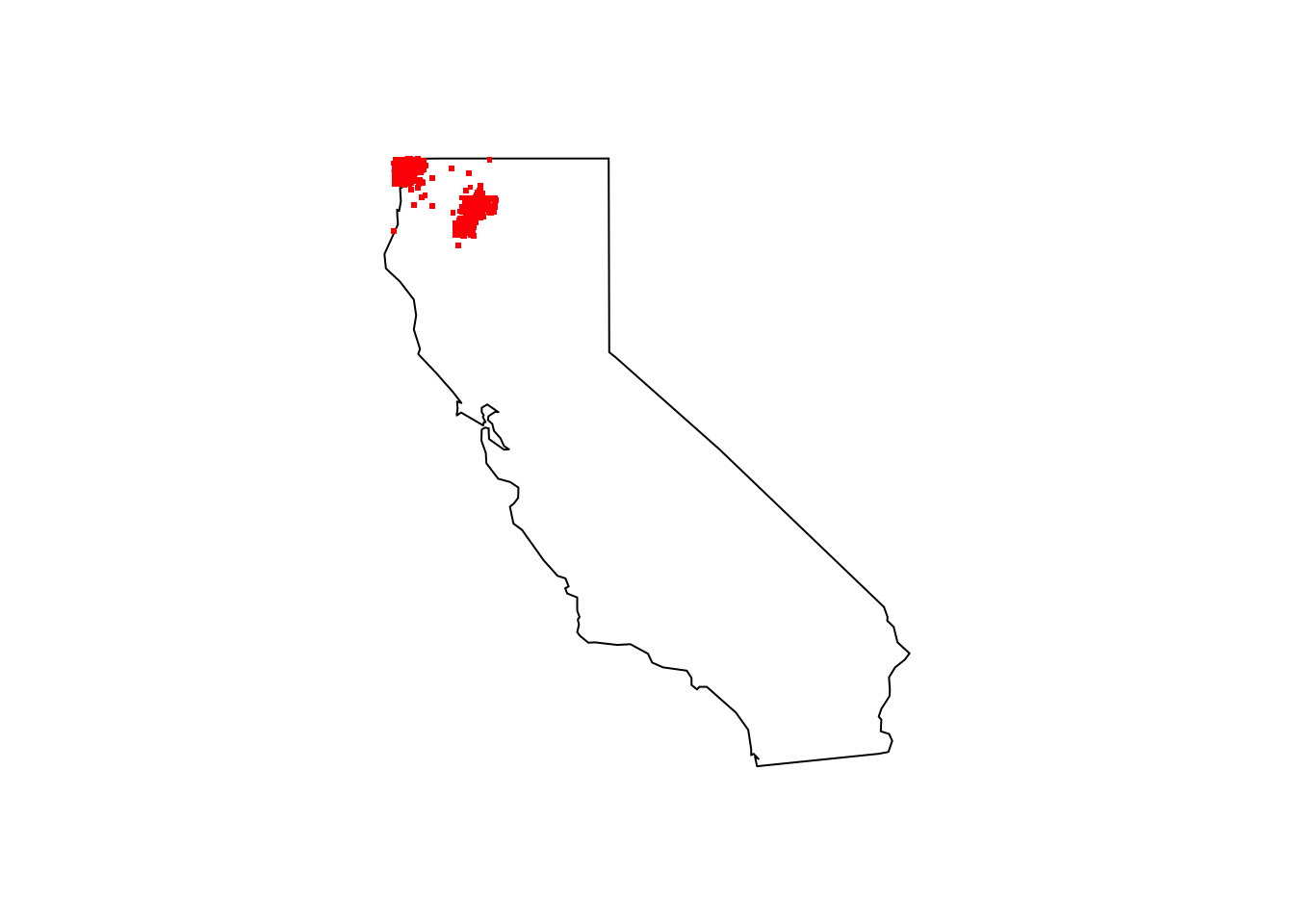
Pull in my run data using the gpx R library and subset the route to make plotting easier.
run <- read_gpx('../../../static/data/cobra_lily_run.gpx')
TrailRun1 <- as.data.frame(run$routes)Make a quick run plot.
TrailRun_plot <- ggplot() +
geom_point(data = TrailRun1, aes(x = Longitude, y = Latitude), color = 'black') +
xlab("Longitude") + ylab("Latitude")
TrailRun_plot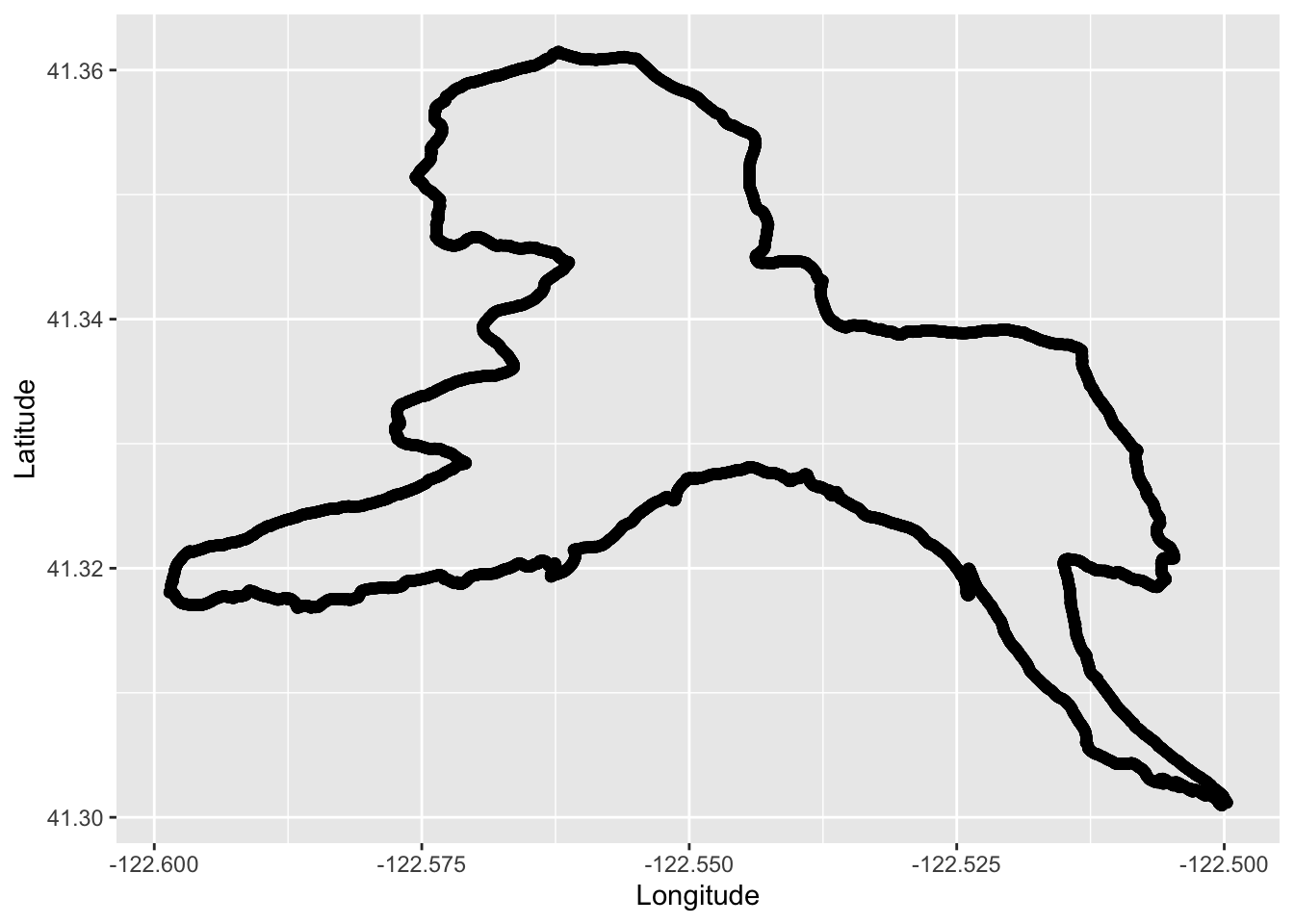
Subset the larger California dataset to only include a zoomed in portion where I ran based on the maximum and minimum coordinates from the run plot above.
cobra_lily_coords_sub <- subset(cobra_lily_coords, decimalLatitude > 41.3 & decimalLatitude < 41.365 & decimalLongitude > -122.6 & decimalLongitude < -122.5 )Overlay the plots and add a triangle for the seep where I observed some Cobra Lilies next to the trail.
TrailRun_plot2 <- ggplot() +
coord_quickmap() +
geom_point(data = TrailRun1, aes(x = Longitude, y = Latitude), color = 'black') +
geom_point(data=cobra_lily_coords_sub, aes(x = decimalLongitude, y = decimalLatitude), color = 'red') +
geom_point(aes(x = -122.55, y = 41.325), pch=25, size= 12, colour="purple") +
xlab("Longitude") + ylab("Latitude")
TrailRun_plot2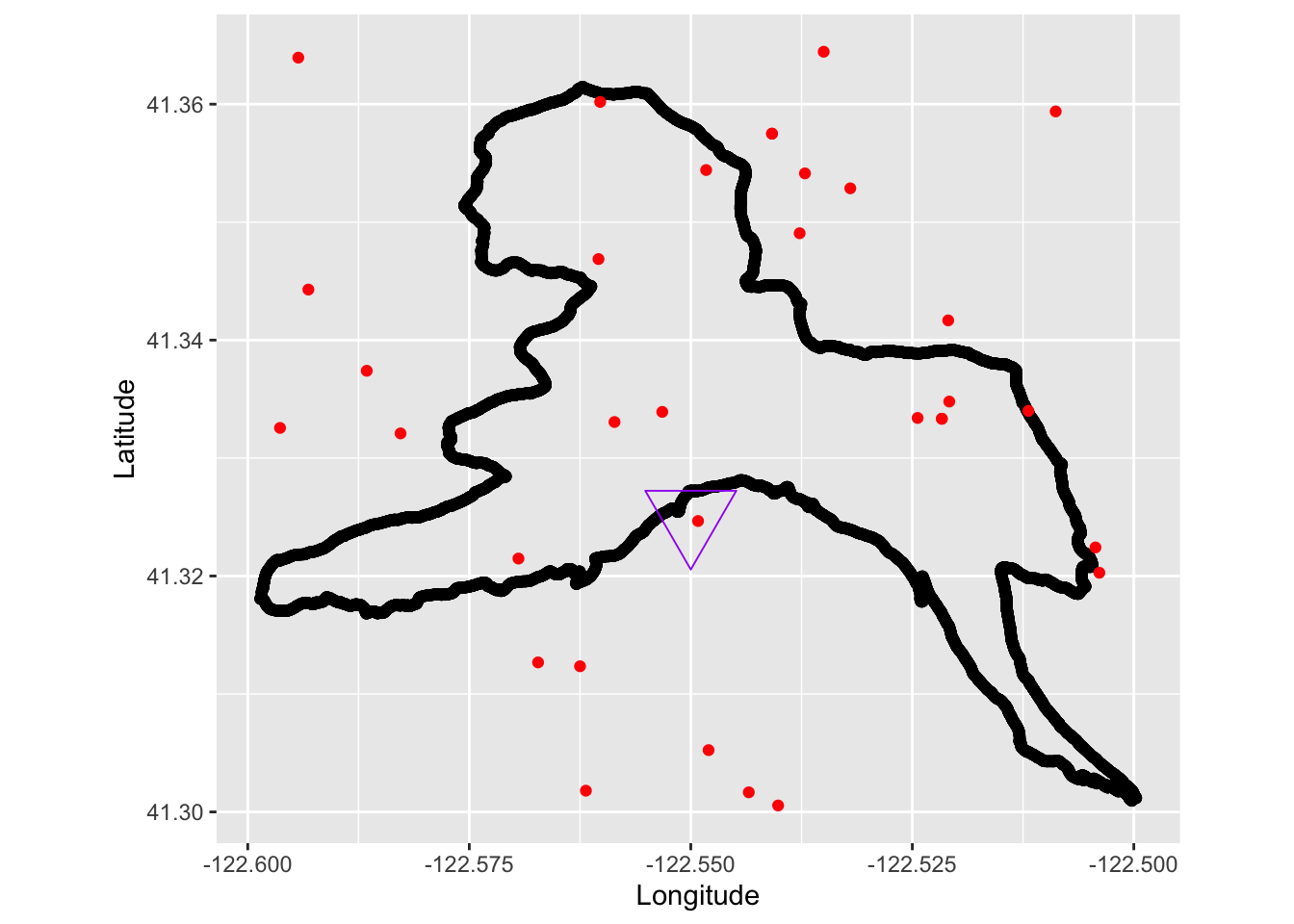
In a follow-up post we will import some additional environmental data to make a multi-layered map.